Multi model comparison#
We often want to compare the result of multiple models.
Calibration. We have several “runs” of the same model with different settings. We would like to find the best.
Validation. We would like to compare our model with alternative models, e.g. a regional DHI model or an external model.
In this notebook, we will consider several wave models for the Southern North Sea and compare to both point measurements and satellite altimetry data.
import numpy as np
import modelskill as ms
Define observations#
o1 = ms.PointObservation('data/SW/HKNA_Hm0.dfs0', item=0, x=4.2420, y=52.6887, name="HKNA")
o2 = ms.PointObservation("data/SW/eur_Hm0.dfs0", item=0, x=3.2760, y=51.9990, name="EPL")
o3 = ms.TrackObservation("data/SW/Alti_c2_Dutch.dfs0", item=3, name="c2")
Define models#
mr1 = ms.model_result('data/SW/HKZN_local_2017_DutchCoast.dfsu', name='SW_1', item=0)
mr2 = ms.model_result('data/SW/HKZN_local_2017_DutchCoast_v2.dfsu', name='SW_2', item=0)
mr3 = ms.model_result('data/SW/ERA5_DutchCoast.nc', name='ERA5', item="swh")
##
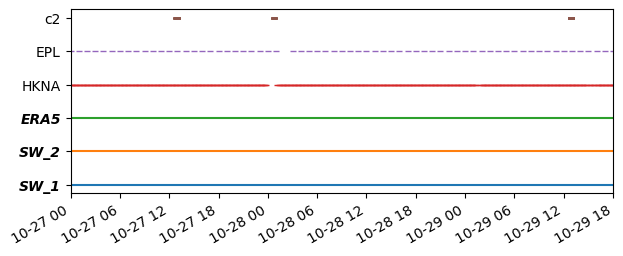
Temporal coverage#
ms.plotting.temporal_coverage([o1, o2, o3], [mr1, mr2, mr3]);
Match observations and model results#
# HKNA is outside ERA5 domain -> pick nearest grid cell
cc = ms.match(obs=[o1, o2, o3], mod=[mr1, mr2, mr3], spatial_method="nearest")
cc
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/mikeio/dataset/_dataset.py:505: FutureWarning: 'inplace' parameter is deprecated and will be removed in future versions. Use ds = ds.rename(...) instead.
warnings.warn(
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/mikeio/dataset/_dataset.py:505: FutureWarning: 'inplace' parameter is deprecated and will be removed in future versions. Use ds = ds.rename(...) instead.
warnings.warn(
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/mikeio/dataset/_dataset.py:505: FutureWarning: 'inplace' parameter is deprecated and will be removed in future versions. Use ds = ds.rename(...) instead.
warnings.warn(
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/mikeio/dataset/_dataset.py:505: FutureWarning: 'inplace' parameter is deprecated and will be removed in future versions. Use ds = ds.rename(...) instead.
warnings.warn(
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/mikeio/dataset/_dataset.py:505: FutureWarning: 'inplace' parameter is deprecated and will be removed in future versions. Use ds = ds.rename(...) instead.
warnings.warn(
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/mikeio/dataset/_dataset.py:505: FutureWarning: 'inplace' parameter is deprecated and will be removed in future versions. Use ds = ds.rename(...) instead.
warnings.warn(
<ComparerCollection>
Comparers:
0: HKNA - Significant wave height [m]
1: EPL - Significant wave height [m]
2: c2 - Significant wave height [m]
cc["EPL"] # select a single comparer from the collection like this
<Comparer>
Quantity: Significant wave height [m]
Observation: EPL, n_points=66
Model(s):
0: SW_1
1: SW_2
2: ERA5
Perform analysis#
You can perform simple filtering on specific observation or specific model. You can refer to observations and models using their name or index.
The main analysis methods are:
skill()
mean_skill()
scatter()
taylor()
cc.skill()
| n | bias | rmse | urmse | mae | cc | si | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| model | observation | ||||||||
| SW_1 | HKNA | 386 | -0.194260 | 0.351964 | 0.293499 | 0.251839 | 0.971194 | 0.094489 | 0.905300 |
| EPL | 66 | -0.066028 | 0.224919 | 0.215009 | 0.189791 | 0.969512 | 0.082773 | 0.932082 | |
| c2 | 102 | -0.025059 | 0.354270 | 0.353383 | 0.297344 | 0.970862 | 0.131869 | 0.889056 | |
| SW_2 | HKNA | 386 | -0.100426 | 0.293033 | 0.275287 | 0.214422 | 0.971194 | 0.088626 | 0.934358 |
| EPL | 66 | 0.001181 | 0.233964 | 0.233961 | 0.199915 | 0.969512 | 0.090069 | 0.926509 | |
| c2 | 102 | 0.050773 | 0.423351 | 0.420296 | 0.353069 | 0.970862 | 0.156839 | 0.841570 | |
| ERA5 | HKNA | 386 | -0.437425 | 0.545329 | 0.325643 | 0.441374 | 0.974863 | 0.104837 | 0.772663 |
| EPL | 66 | -0.207731 | 0.289960 | 0.202299 | 0.223748 | 0.972255 | 0.077880 | 0.887121 | |
| c2 | 102 | -0.390274 | 0.486571 | 0.290582 | 0.407335 | 0.972701 | 0.108435 | 0.790720 |
cc.sel(observation="c2").skill()
| n | bias | rmse | urmse | mae | cc | si | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| model | observation | ||||||||
| SW_1 | c2 | 102 | -0.025059 | 0.354270 | 0.353383 | 0.297344 | 0.970862 | 0.131869 | 0.889056 |
| SW_2 | c2 | 102 | 0.050773 | 0.423351 | 0.420296 | 0.353069 | 0.970862 | 0.156839 | 0.841570 |
| ERA5 | c2 | 102 | -0.390274 | 0.486571 | 0.290582 | 0.407335 | 0.972701 | 0.108435 | 0.790720 |
cc.sel(model=0, observation=[0,"c2"]).mean_skill()
| n | bias | rmse | urmse | mae | cc | si | r2 | |
|---|---|---|---|---|---|---|---|---|
| model | ||||||||
| ERA5 | 488 | -0.413850 | 0.515950 | 0.308112 | 0.424355 | 0.973782 | 0.106636 | 0.781691 |
| SW_1 | 488 | -0.109659 | 0.353117 | 0.323441 | 0.274591 | 0.971028 | 0.113179 | 0.897178 |
| SW_2 | 488 | -0.024826 | 0.358192 | 0.347791 | 0.283745 | 0.971028 | 0.122732 | 0.887964 |
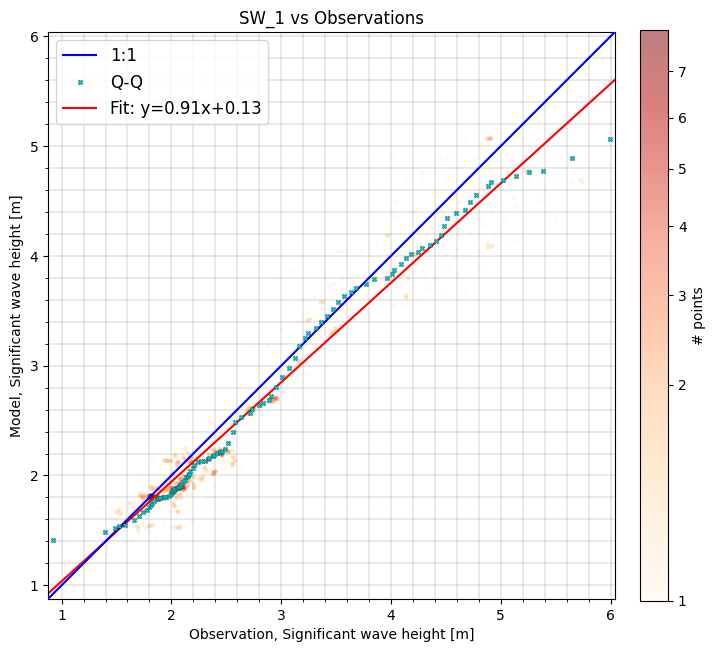
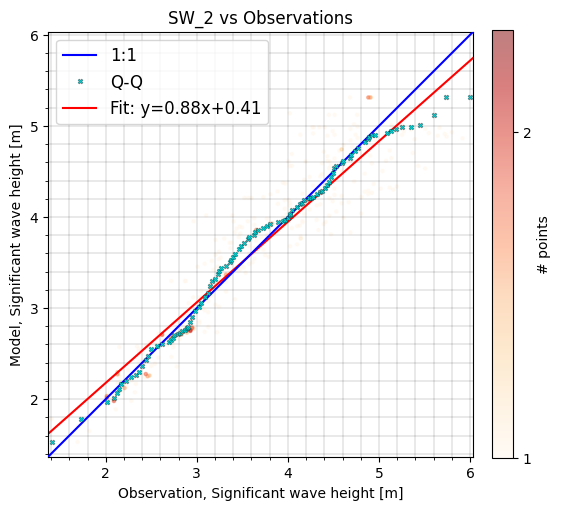
cc.sel(model='SW_1').plot.scatter(cmap='OrRd');
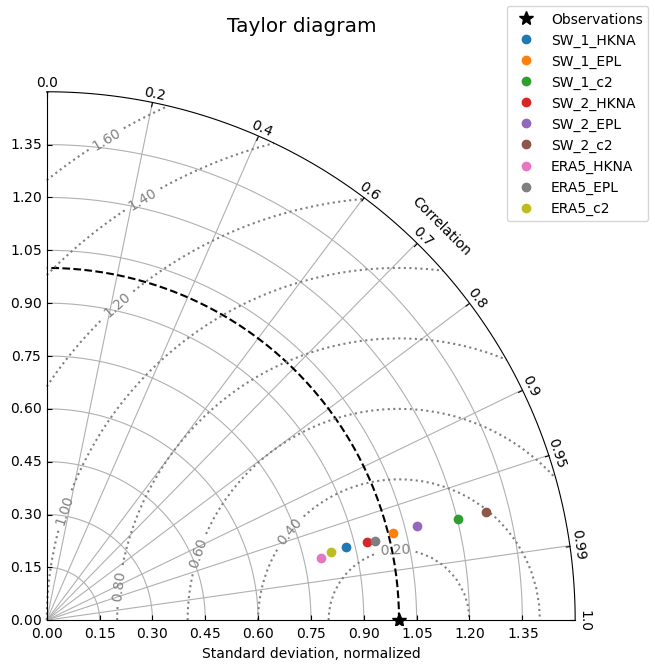
cc.plot.taylor(normalize_std=True, aggregate_observations=False)
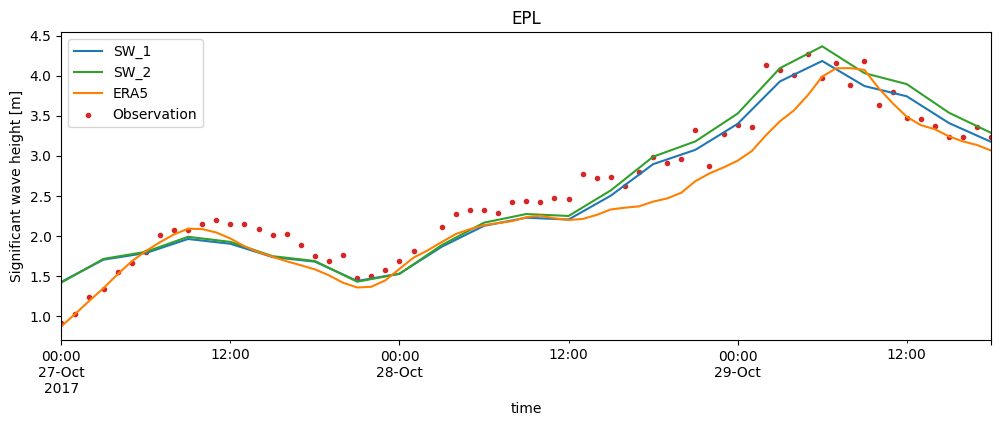
Time series plot (specifically for point comparisons)#
If you select an comparison from the collection which is a PointComparer, you can do a time series plot
cc['EPL'].plot.timeseries(figsize=(12,4));
Filtering on time#
Use the start and end arguments to do your analysis on part of the time series
cc.sel(model="SW_1", end='2017-10-28').skill()
| n | bias | rmse | urmse | mae | cc | si | r2 | |
|---|---|---|---|---|---|---|---|---|
| observation | ||||||||
| HKNA | 281 | -0.097075 | 0.203241 | 0.178559 | 0.164563 | 0.968106 | 0.070167 | 0.916126 |
| EPL | 47 | -0.100029 | 0.240775 | 0.219013 | 0.205827 | 0.919089 | 0.101695 | 0.811727 |
| c2 | 66 | -0.210375 | 0.323525 | 0.245786 | 0.268925 | 0.385374 | 0.121671 | -1.855535 |
cc.sel(model='SW_2', start='2017-10-28').plot.scatter(cmap='OrRd', figsize=(6,7));
Filtering on area#
You can do you analysis in a specific area by providing a bounding box or a closed polygon
bbox = np.array([0.5,52.5,5,54])
polygon = np.array([[6,51],[0,55],[0,51],[6,51]])
cc.sel(model="SW_1", area=bbox).skill()
| n | bias | rmse | urmse | mae | cc | si | r2 | |
|---|---|---|---|---|---|---|---|---|
| observation | ||||||||
| HKNA | 386 | -0.19426 | 0.351964 | 0.293499 | 0.251839 | 0.971194 | 0.094489 | 0.905300 |
| c2 | 42 | -0.05587 | 0.388404 | 0.384365 | 0.336023 | 0.952688 | 0.145724 | 0.749645 |
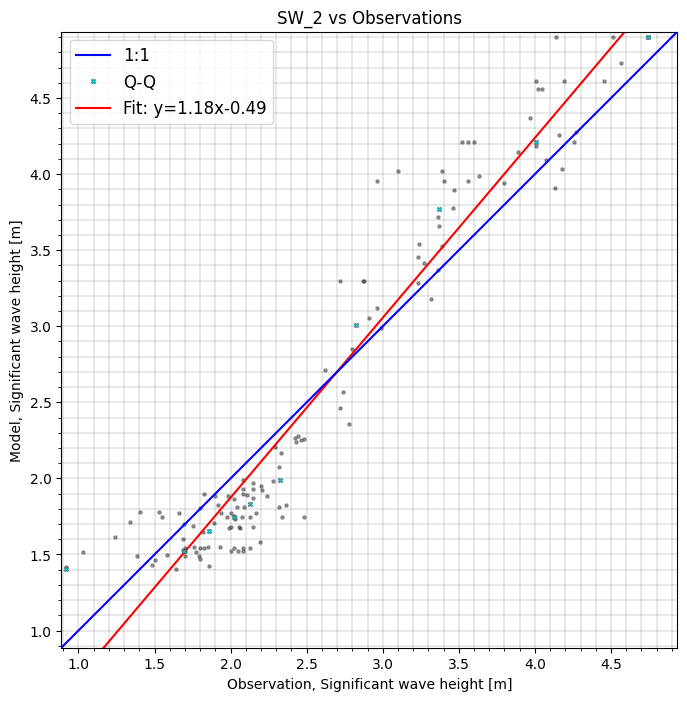
cc.sel(model="SW_2", area=polygon).plot.scatter();
Skill object#
The skill() and mean_skill() methods return a skill object that can visualize results in various ways. The primary methods of the skill object are:
style()
plot_bar()
plot_barh()
plot_line()
plot_grid()
sel()
s = cc.skill()
s.style()
| n | bias | rmse | urmse | mae | cc | si | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| model | observation | ||||||||
| SW_1 | HKNA | 386 | -0.194 | 0.352 | 0.293 | 0.252 | 0.971 | 0.094 | 0.905 |
| EPL | 66 | -0.066 | 0.225 | 0.215 | 0.190 | 0.970 | 0.083 | 0.932 | |
| c2 | 102 | -0.025 | 0.354 | 0.353 | 0.297 | 0.971 | 0.132 | 0.889 | |
| SW_2 | HKNA | 386 | -0.100 | 0.293 | 0.275 | 0.214 | 0.971 | 0.089 | 0.934 |
| EPL | 66 | 0.001 | 0.234 | 0.234 | 0.200 | 0.970 | 0.090 | 0.927 | |
| c2 | 102 | 0.051 | 0.423 | 0.420 | 0.353 | 0.971 | 0.157 | 0.842 | |
| ERA5 | HKNA | 386 | -0.437 | 0.545 | 0.326 | 0.441 | 0.975 | 0.105 | 0.773 |
| EPL | 66 | -0.208 | 0.290 | 0.202 | 0.224 | 0.972 | 0.078 | 0.887 | |
| c2 | 102 | -0.390 | 0.487 | 0.291 | 0.407 | 0.973 | 0.108 | 0.791 |
s.style(columns='rmse')
| n | bias | rmse | urmse | mae | cc | si | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| model | observation | ||||||||
| SW_1 | HKNA | 386 | -0.194 | 0.352 | 0.293 | 0.252 | 0.971 | 0.094 | 0.905 |
| EPL | 66 | -0.066 | 0.225 | 0.215 | 0.190 | 0.970 | 0.083 | 0.932 | |
| c2 | 102 | -0.025 | 0.354 | 0.353 | 0.297 | 0.971 | 0.132 | 0.889 | |
| SW_2 | HKNA | 386 | -0.100 | 0.293 | 0.275 | 0.214 | 0.971 | 0.089 | 0.934 |
| EPL | 66 | 0.001 | 0.234 | 0.234 | 0.200 | 0.970 | 0.090 | 0.927 | |
| c2 | 102 | 0.051 | 0.423 | 0.420 | 0.353 | 0.971 | 0.157 | 0.842 | |
| ERA5 | HKNA | 386 | -0.437 | 0.545 | 0.326 | 0.441 | 0.975 | 0.105 | 0.773 |
| EPL | 66 | -0.208 | 0.290 | 0.202 | 0.224 | 0.972 | 0.078 | 0.887 | |
| c2 | 102 | -0.390 | 0.487 | 0.291 | 0.407 | 0.973 | 0.108 | 0.791 |
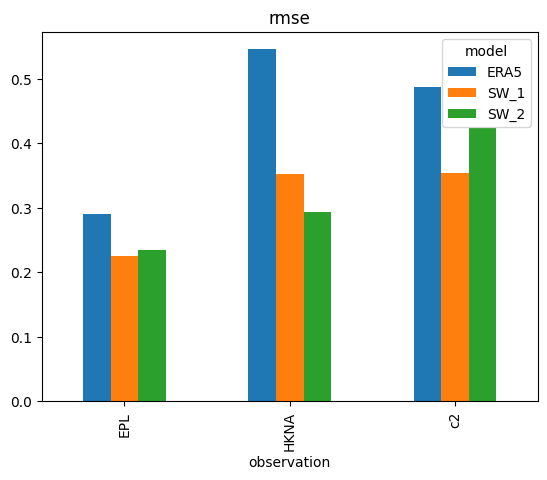
s['rmse'].plot.bar();
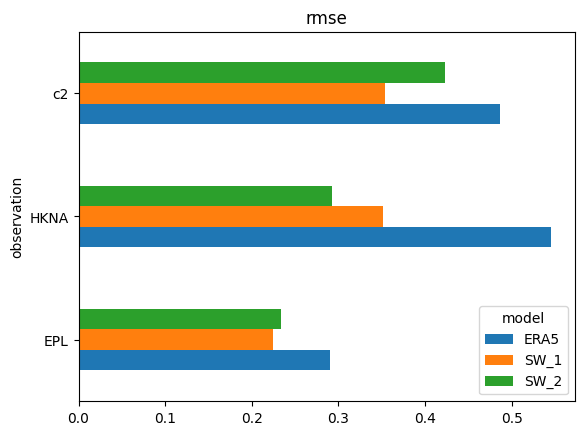
s['rmse'].plot.barh(); # horizontal version
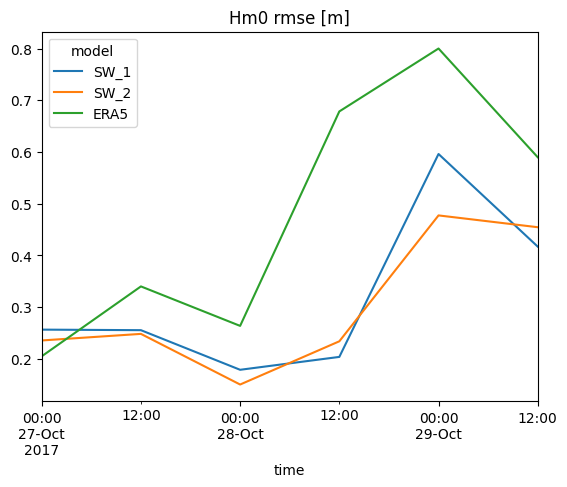
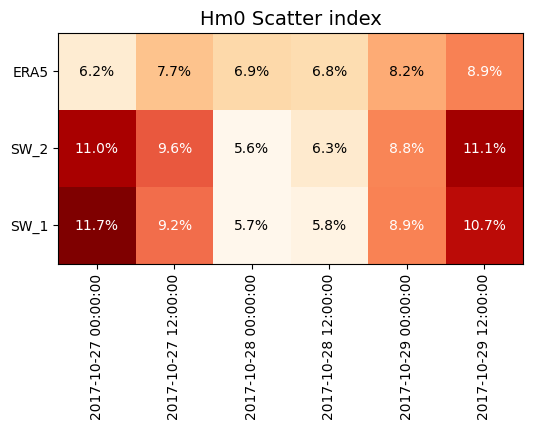
s = cc.skill(by=['model','freq:12H'], metrics=['bias','rmse','si'])
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/modelskill/comparison/_utils.py:153: FutureWarning: 'H' is deprecated and will be removed in a future version, please use 'h' instead.
res.append(pd.Grouper(key="time", freq=freq))
s.style()
| n | bias | rmse | si | ||
|---|---|---|---|---|---|
| model | time | ||||
| SW_1 | 2017-10-27 00:00:00 | 84 | -0.096 | 0.257 | 0.117 |
| 2017-10-27 12:00:00 | 150 | -0.175 | 0.255 | 0.092 | |
| 2017-10-28 00:00:00 | 76 | -0.111 | 0.179 | 0.057 | |
| 2017-10-28 12:00:00 | 84 | -0.037 | 0.204 | 0.058 | |
| 2017-10-29 00:00:00 | 82 | -0.421 | 0.596 | 0.089 | |
| 2017-10-29 12:00:00 | 78 | -0.020 | 0.417 | 0.107 | |
| SW_2 | 2017-10-27 00:00:00 | 84 | -0.070 | 0.236 | 0.110 |
| 2017-10-27 12:00:00 | 150 | -0.157 | 0.248 | 0.096 | |
| 2017-10-28 00:00:00 | 76 | -0.055 | 0.150 | 0.056 | |
| 2017-10-28 12:00:00 | 84 | 0.091 | 0.234 | 0.063 | |
| 2017-10-29 00:00:00 | 82 | -0.226 | 0.477 | 0.088 | |
| 2017-10-29 12:00:00 | 78 | 0.141 | 0.455 | 0.111 | |
| ERA5 | 2017-10-27 00:00:00 | 84 | -0.161 | 0.205 | 0.062 |
| 2017-10-27 12:00:00 | 150 | -0.303 | 0.340 | 0.077 | |
| 2017-10-28 00:00:00 | 76 | -0.200 | 0.264 | 0.069 | |
| 2017-10-28 12:00:00 | 84 | -0.637 | 0.679 | 0.068 | |
| 2017-10-29 00:00:00 | 82 | -0.699 | 0.800 | 0.082 | |
| 2017-10-29 12:00:00 | 78 | -0.479 | 0.590 | 0.089 |
s['rmse'].plot.line(title='Hm0 rmse [m]');
s.plot.grid('si', fmt='0.1%', title='Hm0 Scatter index');
/opt/hostedtoolcache/Python/3.13.11/x64/lib/python3.13/site-packages/modelskill/skill.py:290: FutureWarning: Selecting metric in plot functions like modelskill.skill().plot.grid(si) is deprecated and will be removed in a future version. Use modelskill.skill()['si'].plot.grid() instead.
warnings.warn(
The sel() method can subset the skill object#
A new skill object will be returned
s = cc.skill()
s.style()
| n | bias | rmse | urmse | mae | cc | si | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| model | observation | ||||||||
| SW_1 | HKNA | 386 | -0.194 | 0.352 | 0.293 | 0.252 | 0.971 | 0.094 | 0.905 |
| EPL | 66 | -0.066 | 0.225 | 0.215 | 0.190 | 0.970 | 0.083 | 0.932 | |
| c2 | 102 | -0.025 | 0.354 | 0.353 | 0.297 | 0.971 | 0.132 | 0.889 | |
| SW_2 | HKNA | 386 | -0.100 | 0.293 | 0.275 | 0.214 | 0.971 | 0.089 | 0.934 |
| EPL | 66 | 0.001 | 0.234 | 0.234 | 0.200 | 0.970 | 0.090 | 0.927 | |
| c2 | 102 | 0.051 | 0.423 | 0.420 | 0.353 | 0.971 | 0.157 | 0.842 | |
| ERA5 | HKNA | 386 | -0.437 | 0.545 | 0.326 | 0.441 | 0.975 | 0.105 | 0.773 |
| EPL | 66 | -0.208 | 0.290 | 0.202 | 0.224 | 0.972 | 0.078 | 0.887 | |
| c2 | 102 | -0.390 | 0.487 | 0.291 | 0.407 | 0.973 | 0.108 | 0.791 |
s.sel(model='SW_1').style()
| model | n | bias | rmse | urmse | mae | cc | si | r2 | |
|---|---|---|---|---|---|---|---|---|---|
| observation | |||||||||
| HKNA | SW_1 | 386 | -0.194 | 0.352 | 0.293 | 0.252 | 0.971 | 0.094 | 0.905 |
| EPL | SW_1 | 66 | -0.066 | 0.225 | 0.215 | 0.190 | 0.970 | 0.083 | 0.932 |
| c2 | SW_1 | 102 | -0.025 | 0.354 | 0.353 | 0.297 | 0.971 | 0.132 | 0.889 |
s.sel(observation='HKNA').style()
| observation | n | bias | rmse | urmse | mae | cc | si | r2 | |
|---|---|---|---|---|---|---|---|---|---|
| model | |||||||||
| SW_1 | HKNA | 386 | -0.194 | 0.352 | 0.293 | 0.252 | 0.971 | 0.094 | 0.905 |
| SW_2 | HKNA | 386 | -0.100 | 0.293 | 0.275 | 0.214 | 0.971 | 0.089 | 0.934 |
| ERA5 | HKNA | 386 | -0.437 | 0.545 | 0.326 | 0.441 | 0.975 | 0.105 | 0.773 |
s.query('rmse>0.25').style()
| n | bias | rmse | urmse | mae | cc | si | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| model | observation | ||||||||
| SW_1 | HKNA | 386 | -0.194 | 0.352 | 0.293 | 0.252 | 0.971 | 0.094 | 0.905 |
| c2 | 102 | -0.025 | 0.354 | 0.353 | 0.297 | 0.971 | 0.132 | 0.889 | |
| SW_2 | HKNA | 386 | -0.100 | 0.293 | 0.275 | 0.214 | 0.971 | 0.089 | 0.934 |
| c2 | 102 | 0.051 | 0.423 | 0.420 | 0.353 | 0.971 | 0.157 | 0.842 | |
| ERA5 | HKNA | 386 | -0.437 | 0.545 | 0.326 | 0.441 | 0.975 | 0.105 | 0.773 |
| EPL | 66 | -0.208 | 0.290 | 0.202 | 0.224 | 0.972 | 0.078 | 0.887 | |
| c2 | 102 | -0.390 | 0.487 | 0.291 | 0.407 | 0.973 | 0.108 | 0.791 |

