Dfs1#
Dfs1 has in addition to the dfs0 file a single spatial dimension.
This spatial dimension has information on the grid spacing, but do not contain enough metadata to determine their geographical position, but have a relative distance from the origo.
See Dfs1 in MIKE IO Documentation
import mikeio
ds = mikeio.read("data/waterlevel_north.dfs1")
ds
<mikeio.Dataset>
dims: (time:577, x:2)
time: 1993-12-02 00:00:00 - 1993-12-14 00:00:00 (577 records)
geometry: Grid1D (n=2, dx=8800)
items:
0: North WL <Water Level> (meter)
ds.geometry
<mikeio.Grid1D>
x: [0, 8800] (nx=2, dx=8800)
This dfs1 file contains only two nodes (nx=2) and has a grid spacing of 8800 m.
The dataset has a single item North WL which we can access in three different ways.
As an attribute
ds.North_WLAs a key in a dictionary
ds["North WL"]By position
ds[0]
da = ds.North_WL
da
<mikeio.DataArray>
name: North WL
dims: (time:577, x:2)
time: 1993-12-02 00:00:00 - 1993-12-14 00:00:00 (577 records)
geometry: Grid1D (n=2, dx=8800)
The dataarray variable da contains all the data for the North WL item from the dfs1 file.
Visualization#
The data can be visualized with the .plot() method.
The default plot show time on the vertical axis and space on the horizontal axis since dfs1 are often used to represent horizontal variations along an open boundary.
da.plot();
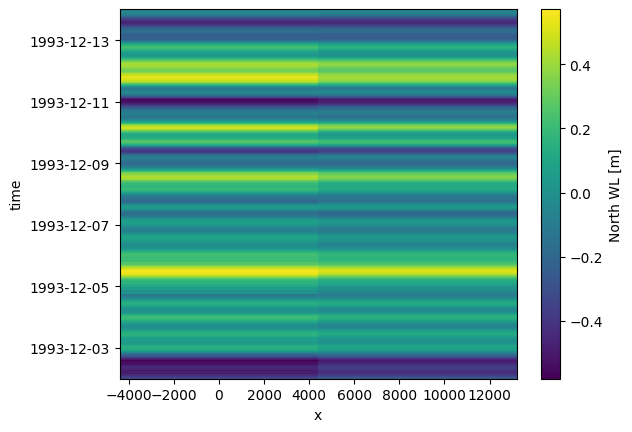
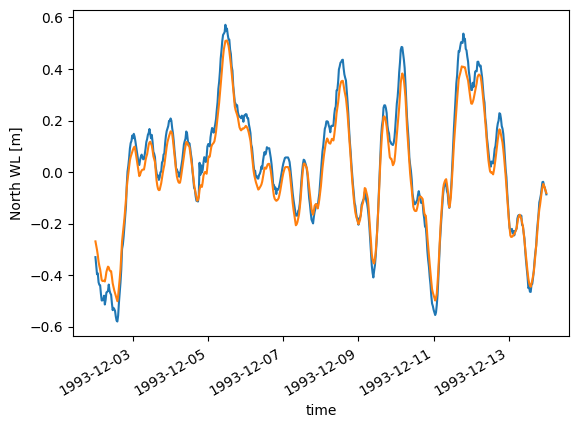
Fort this example, with only a few nodes on the spatial axis, it makes more sense to visualize the data as individual lines in a timeseries plot.
da.plot.timeseries();
Inline exercise
Are there other possible visualizations?
Spatial and temporal selection#
Spatial subsetting can be done in two ways:
positional index using
.isel()x=0..(nx-1) (or with negative indexing from the end)distance along the axis using
sel()x=0..8800 (pick closest node)
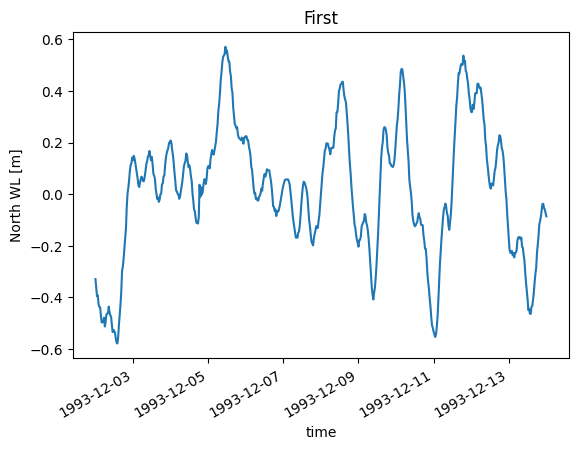
da.isel(x=0).plot(title="First");
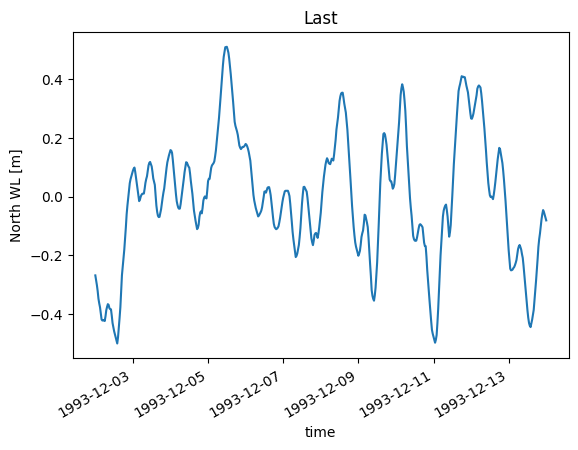
da.isel(x=-1).plot(title="Last");
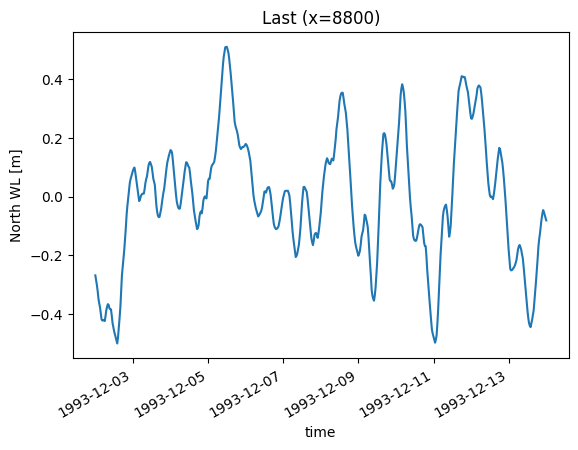
da.sel(x=8800).plot(title="Last (x=8800)")
<Axes: title={'center': 'Last (x=8800)'}, xlabel='time', ylabel='North WL [m]'>
Calculated items#
Creating a new dataarray in the dataset based on an existing dataarray is easy.
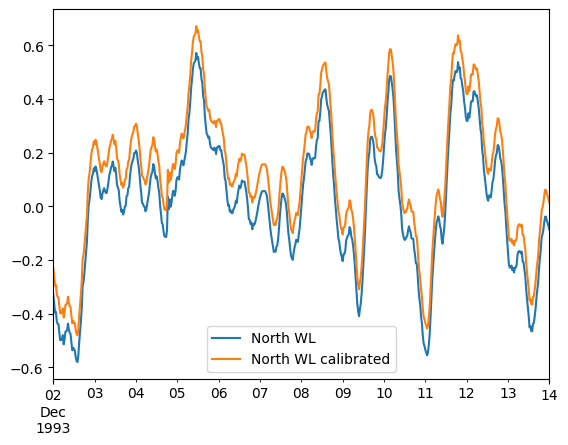
ds["North WL calibrated"] = da + 0.1 # add 0.1 m to the original data
Export to dfs0#
After subsetting into a single spatial point the result no longer has any spatial dimension and has become a simple timeseries which can be saved as a dfs0 file.
ds.isel(x=0)
<mikeio.Dataset>
dims: (time:577)
time: 1993-12-02 00:00:00 - 1993-12-14 00:00:00 (577 records)
geometry: GeometryUndefined()
items:
0: North WL <Water Level> (meter)
1: North WL calibrated <Water Level> (meter)
ds.isel(x=0).geometry
GeometryUndefined()
ds.isel(x=0).to_dfs("random_0.dfs0")
mikeio.read("random_0.dfs0").plot();
Inline exercise
Export the timeseries in the last spatial element as a csv file
import utils
utils.sysinfo()
System: 3.11.14 (main, Oct 10 2025, 01:03:14) [GCC 13.3.0]
NumPy: 2.4.2
Pandas: 3.0.0
MIKE IO: 3.0.1
Last modified: 2026-02-10 13:11:59.630807

