import matplotlib.pyplot as plt
import mikeioDfs3 - Basic
ds = mikeio.read("../tests/testdata/dissolved_oxygen.dfs3")
ds<mikeio.Dataset>
dims: (time:1, z:17, y:112, x:91)
time: 2001-12-28 00:00:00 (time-invariant)
geometry: Grid3D(nz=17, ny=112, nx=91)
items:
0: Diss. oxygen (mg/l) <Concentration 3> (mg per liter)ds.geometry<mikeio.Grid3D>
x: [0, 150, ..., 1.35e+04] (nx=91, dx=150)
y: [0, 150, ..., 1.665e+04] (ny=112, dy=150)
z: [0, 1, ..., 16] (nz=17, dz=1)
origin: (10.37, 55.42), orientation: 18.125
projection: PROJCS["UTM-32",GEOGCS["Unused",DATUM["UTM Projections",SPHEROID["WGS 1984",6378137,298.257223563]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Transverse_Mercator"],PARAMETER["False_Easting",500000],PARAMETER["False_Northing",0],PARAMETER["Central_Meridian",9],PARAMETER["Scale_Factor",0.9996],PARAMETER["Latitude_Of_Origin",0],UNIT["Meter",1]]do = ds[0]
do<mikeio.DataArray>
name: Diss. oxygen (mg/l)
dims: (time:1, z:17, y:112, x:91)
time: 2001-12-28 00:00:00 (time-invariant)
geometry: Grid3D(nz=17, ny=112, nx=91)do.isel(z=-1).plot();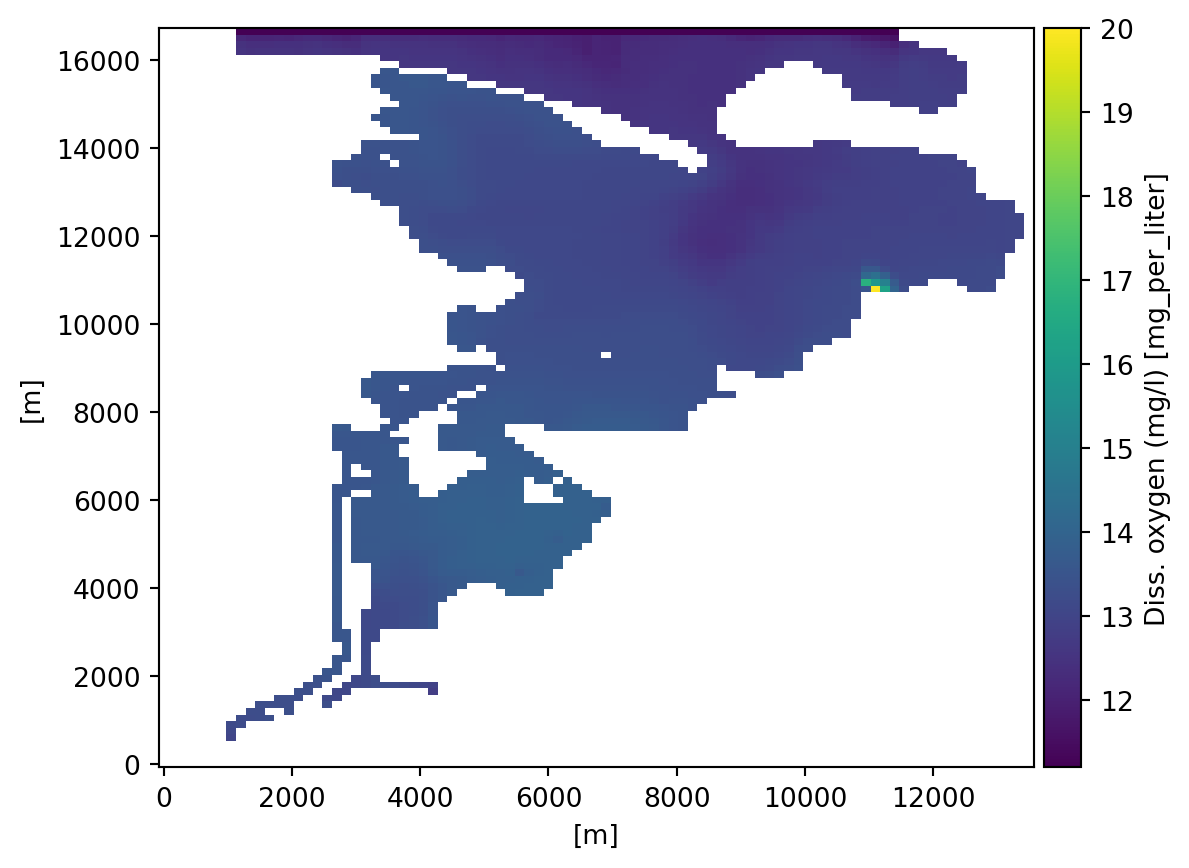
dst = mikeio.read("../tests/testdata/dissolved_oxygen.dfs3", layers="top")dst<mikeio.Dataset>
dims: (time:1, y:112, x:91)
time: 2001-12-28 00:00:00 (time-invariant)
geometry: Grid2D (ny=112, nx=91)
items:
0: Diss. oxygen (mg/l) <Concentration 3> (mg per liter)dst[0].plot();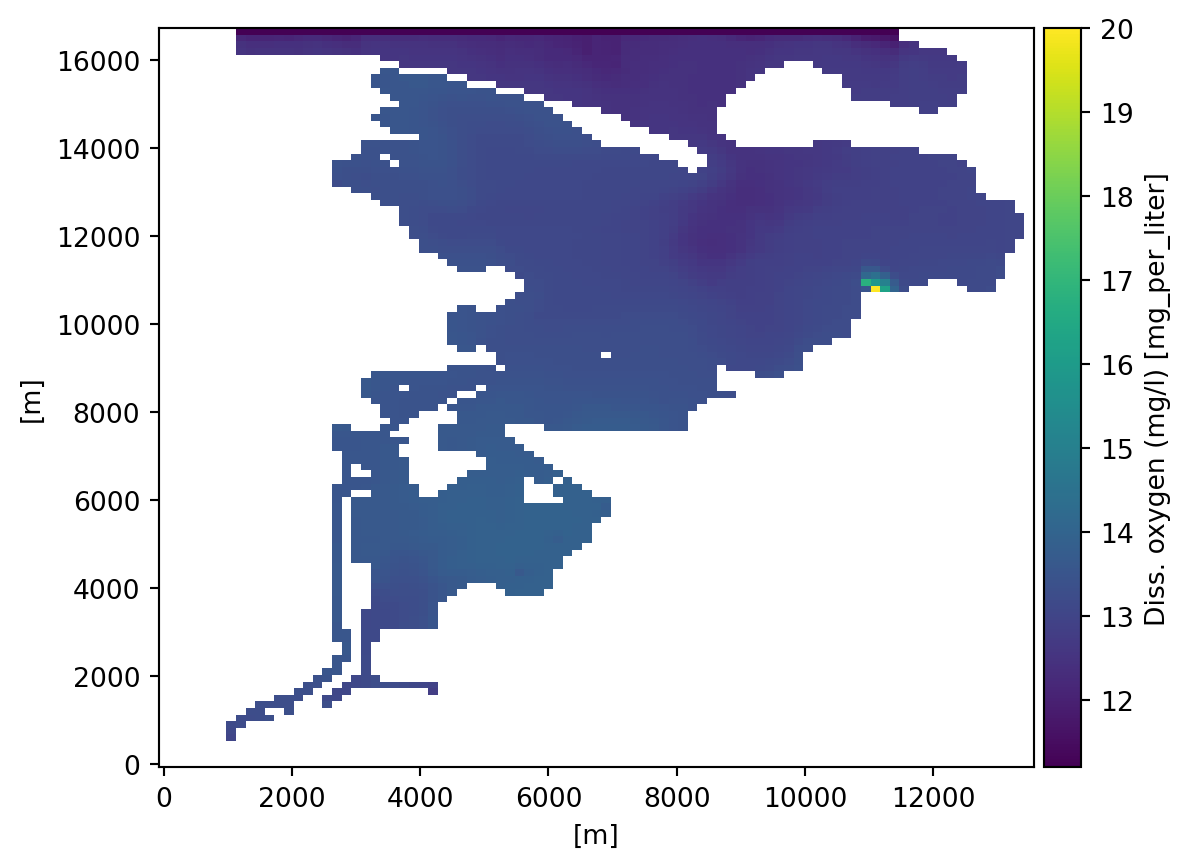
dsb = mikeio.read("../tests/testdata/dissolved_oxygen.dfs3",layers="bottom")
dsb<mikeio.Dataset>
dims: (time:1, y:112, x:91)
time: 2001-12-28 00:00:00 (time-invariant)
geometry: Grid2D (ny=112, nx=91)
items:
0: Diss. oxygen (mg/l) <Concentration 3> (mg per liter)dsb[0].plot(figsize=(10,10))
plt.title("Bottom oxygen");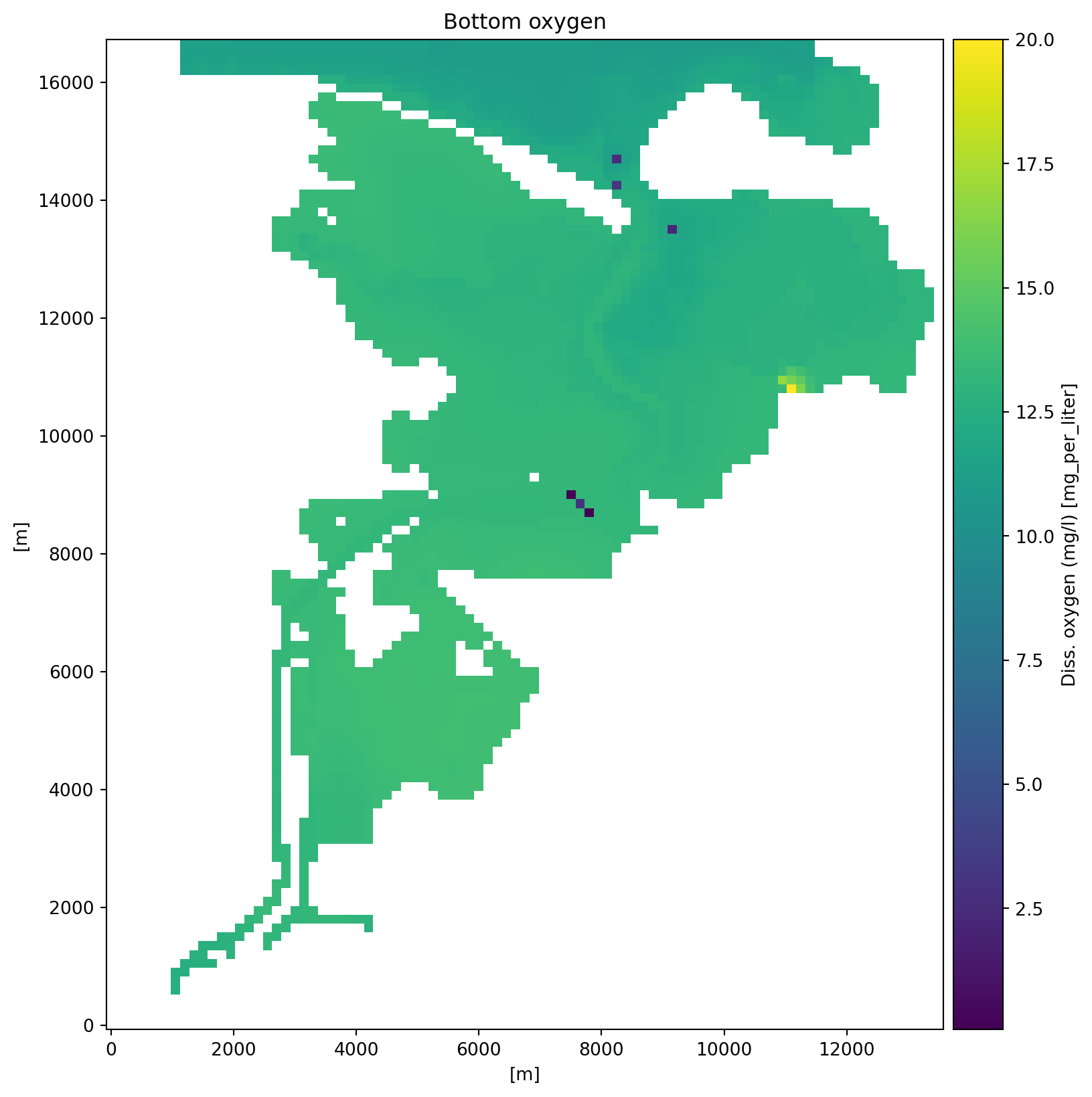
dsb[0].to_numpy()[0,110,56]11.076560020446777dst[0].to_numpy()[0,110,56]12.409002304077148dsb[0].to_numpy()[0,58,52]0.05738005042076111